Researchers at KEK solved the crystal structure of a protein complex between NEMO (NF-κB essential modulator) and linear polyubiquitin chain. The interaction between NEMO and linear polyubiquitn chain is crucial for NF-κB (nuclear factor kappa B) activation which is related to various diseases such as cancer, inflammation and immunodeficiency. The molecular mechanism of interaction would contribute towards structure-based drug design against NF-κB activation.
Researchers at Structural Biology Research Center, Institute of Materials Structure Science, KEK, led by director Soichi Wakatsuki, in collaboration with researchers at Goethe University medical school, Germany, led by Prof. Ivan Dikic and at Medical Research Council, UK, have succeeded in crystallization of NEMO (NF-κB essential modulator) protein in complex with linear polyubiquitin chains where ubiquitin proteins are connected in a head-to-tail manner. They solved the crystal structure of the complex using beamline BL-17A of Photon Factory (PF), KEK. NEMO is a regulatory subunit of IKK (IκB kinase) complex which phosphorylates IκB (inhibitor of NF-κB) and activates the transcription factor NF-κB (nuclear factor kappa B) which is involved in various diseases such as cancer, inflammation and immunodeficiency. Linear polyubiquitin chains play an important role in activation of NF-κB, through their binding to NEMO. The molecular basis of their interaction would enable us to understand DNA transcription and to develop therapy targeting of NF-κB.
This work will be published in the March 20, 2009 issue of the scientific journal "Cell" published by Cell Press, USA.
NF-κB exists in all animal cells including invertebrates, and activates transcription of various genes involved in inflammation, cell growth, apoptosis and so on. In a resting state of cells, NF-κB stays in cytosol in an inactivated form by binding of its inhibitor IκB (inhibitor of NF-κB). Upon extracellular stimulation, IKK (IκB kinase) complex is activated and phospholyrates IκB. IκB is subsequently polyubiquitinated and degraded by the proteasome. Then free NF-κB enters nucleus and acts as a transcription factor to regulate expression of many genes which are necessary for inflammatory response and so on (Fig. 1). Dysfunction of NF-κB is therefore cause cancer, immunodeficiency and inflammatory diseases.
Ubiquitin is ubiquitously expressed in all eukaryotic cells. Ubiquitin consists of only 76 amino acids, but functions as a versatile signal by binding to target proteins in cells. Ubiquitin modifies proteins by forming isopeptide bond between the last 76th (carboxy terminal) glycine of ubiquitin and lysine side chain on the target proteins. Ubiquitin itself contains seven lysine residues. Then ubiquitin forms chains by isopeptide bond between the carboxy terminal glycine of the second ubiquitin (called distal ubiquitin) and one of the seven lysine residues of the first ubiquitin (called proximal ubiquitin). Multiple ubiquitins can be connected to form polyubiquitin chains. The proteasome selectively degrade proteins which are tagged by Lys48-linked polyubiqutin chains. The work leading to the discovery of this ubiquitin-proteasome system was awarded 2004 Nobel prize. Now it is known that different linkage of polyubiquitin chains are used as various signals in cells not only for protein degradation.
Recently a novel linear polyubiquitin chain modification was discovered by Prof. Kazuhiro Iwai of Osaka University. Linear polyubiquitin chain is formed by normal polypeptide linkage between the carboxy terminal glycine and amino terminal methionine, not between glycine and lysine side chains. Prof. Iwai's group revealed that linear polyubiquitin modification is crucial to NF-κB activation. However, molecular mechanism of linear polyubiquitin recognition by NEMO remained unknown.
The research group at KEK first crystallized ubiquitin binding domain called UBAN (or CoZi) of NEMO and found NEMO forms a long coiled-coil homodimer. They then crystallized NEMO in complex with two tandem ubiquitins as a minimum unit of linear polyubiquitn chains. All the data for structure determination was collected at beamline BL-17 at Photon Factory, KEK.
The structures revealed three important points.
- NEMO makes one long extended α-helix structure. Two NEMO helices coiled together to build up super helical structure called coiled-coil. NEMO dimer shows nearly 2-fold symmetry except for a kink in the middle of coiled-coil (Fig. 2).
- Two linear polyubiquitin chains bind to both sides of the coiled-coil NEMO dimer (Fig. 3). The distal ubiquitin and proximal ubiquitin show different modes of interaction with NEMO.
- Linkage between distal and proximal ubiquitins is extensively recognized by NEMO and therefore polyubiquitins with other linkage are not able to bind in the same way as linear polyubiquitin thus enabling selectivity of NEMO for linkage types of polyubiquitin chains.
Mutational analyses based on the complex structure confirmed that both distal and proximal ubiquitin bindings are necessary for full NF-κB activation. Importantly, mutations found in patients of anhidrotic ectodermal dysplasia with immunodeficiency occur at the NEMO residues which recognize linkage of linear polyubiquitin chains.
Their findings revealed how linear polyubiquitin chains bind to NEMO in the activation cascade of NF-κB. These would provide us with an understanding of polyubiqutin chain selectivity and useful insights into developing therapy targeting of NF-κB.
| [ Related Web Site ] |
Photon Factory |
| [ Media Contact ] |
Soichi Wakatsuki, KEK Insititue of Materials Structure Science
+81 29-864-5631
Youhei Morita, KEK Public Relations Office
+81 29-879-6047
|
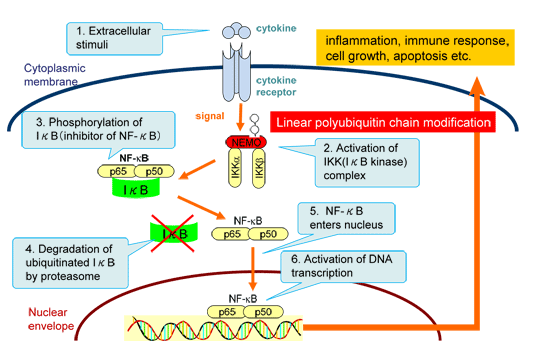 |
Figure 1 : NF-κB activation cascade in cells upon extracellular stimulation. |
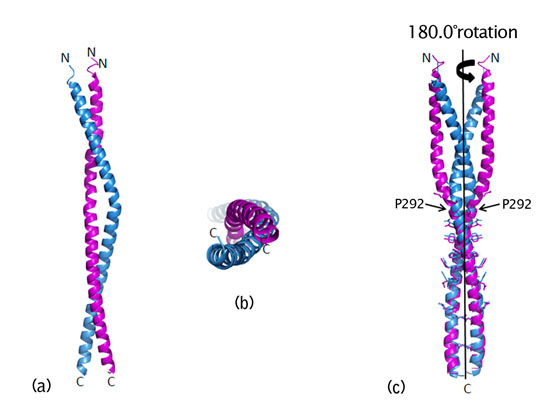 |
Figure 2 : Crystal structure of the coiled-coil dimer of NEMO CoZi domain. (a) Side view of the coiled-coil. (b) Top view of the coiled-coil. (c) Superimposition of the structures rotated by 180 degrees. |
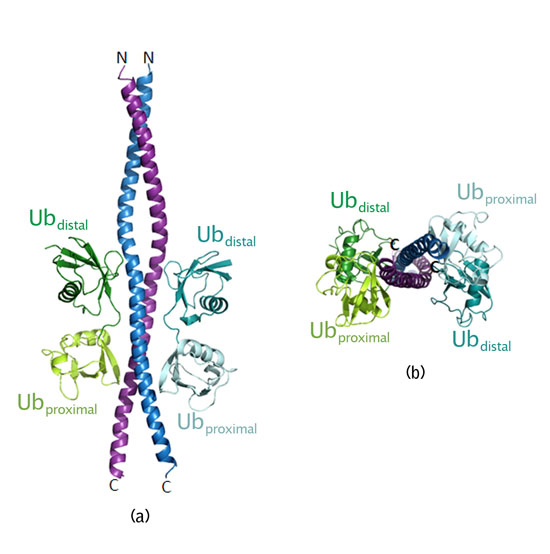 |
Figure 3 : Crystal structure of NEMO dimer in complex with two linear polyubiquitin chains. Each polyubiquitin chain consists of distal and proximal ubiquitins (a) Side view (b) Top view. |
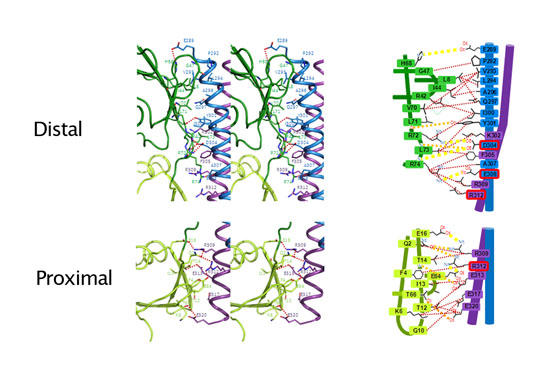 |
Figure 4 : Stereoview (left) and schematic drawing (right) of the interface between NEMO and linear polyubiquitin chains. Different surfaces of ubiquitin are used for distal and proximal interactions with NEMO. These interactions with NEMO are possible only with linear polyubiquitin chains but not with other linkages. |
|



